About
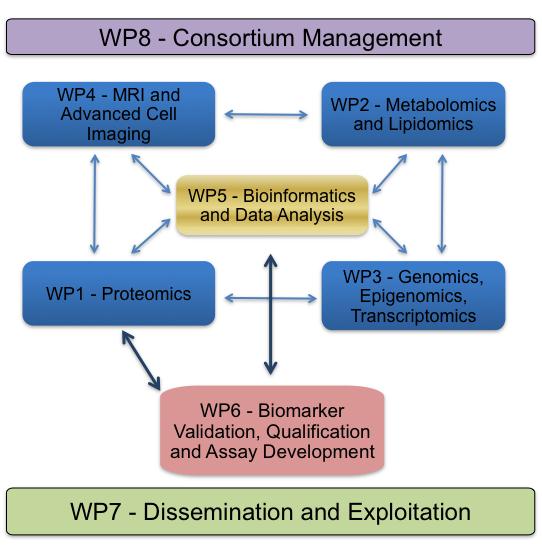
The D-BOARD consortium will use high-throughput genomic tools to identify the genes that increase susceptibility to OA and genetic, epigenetic, genomic and transcriptomic strategies (WP3) to explore the gene regulatory mechanisms involved. Proteomics (WP1), metabolomics and lipidomics (WP2) will be used to identify panels of proteins, glycoproteins and lipid biomarkers that can facilitate the early diagnosis of arthritis in body fluids of human subjects, animal models and in vitro culture systems. Imaging and live cell tracking (WP4) will be used to assess the presence, infiltration and biological contribution of inflammatory cells in animal models of experimental OA and in human OA patients.
Bioinformatics, data modelling and statistical techniques (WP5) will be used to determine the predictive power of the panels of combination biomarkers. Molecular, biochemical and immunologic approaches (WP6) will be used to develop and test multiplex diagnostic and prognostic assays for combinations of cellular, protein, glycoprotein and lipid biomarkers. We will integrate the information obtained to establish mechanisms of disease initiation as well as identification of new genes, proteins and cellular pathways as therapeutic targets and more effective pipelines for the development of safer pharmaceuticals and other treatments.