Contact
Biography
B.Sc. University of Wales (1990); Ph.D. University of Edinburgh (1997); Postdoctoral Research Fellow, University of Nottingham (1996-1999); Research Fellow, The Natural History Museum, London (1999-2001); Lecturer, University of Nottingham (2001-present); Visiting Professor, Chulalongkorn University, Bangkok (2010-present); Fellow of the Linnean Society of London (2022-present).
Google Scholar: https://scholar.google.co.uk/citations?hl=en&user=nDyT5GsAAAAJ&view_op=list_works&sortby=pubdate
ResearchGate: https://www.researchgate.net/profile/Christopher-Wade-3/research
Orcid: https://orcid.org/0000-0002-1269-9694
Teaching Summary
LIFE4141 Comparative and Evolutionary Genomics (lecturer), LIFE4051 Research Presentation Skills (lecturer) LIFE3022 Year 3 Research Project (supervisor), LIFE3041 Molecular Evolution (convenor),… read more
Research Summary
My lab uses DNA sequences to answer questions in parasitology, evolutionary biology, taxonomy and biogeography. Our research falls in 4 main areas:
1) Medical and Veterinary Parasitology: The role of snails as intermediate hosts for medically and veterinary important parasitic diseases (including Schistosomiasis, Angiostrongyliasis and Fascioliasis).
2) Molecular Phylogenetics: Evolutionary relationships and taxonomy of terrestrial and aquatic snails and slugs.
3) Marine Evolution and Ecology: Mapping the distribution of genetic variation in marine taxa, focusing on foraminifera and fish.
4) Bioinformatics.
1) Medical and Veterinary Parasitology: The Role of Snails as Intermediate Hosts for Medically and Veterinary Important Parasitic Diseases (including Schistosomiasis, Angiostrongyliasis and Fascioliasis).
Aquatic snails act as intermediate hosts for trematodes including both Schistosoma and Fasciola. Schistosoma causes schistosomiasis (bilharzia) in humans and our research focuses on screening Biomphalaria and Bulinus snails for schistosome infection in order to determine the incidence of schistosomiasis and the location of sites with active transmission. We also examine population genetic variation in host snail species in order to better understand the link between host snail genetic diversity and infection. Our research on schistosomiasis is centred on African species and we are currently undertaking active research in The Gambia, Nigeria and East Africa (Kenya, Tanzania and Uganda). Fasciola (liver fluke) causes disease in humans and animals the world over and we use molecular techniques to monitor aquatic snails species for Fasciola infection.
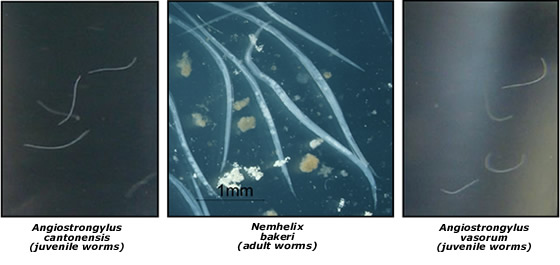
Snails and slugs also act as intermediate hosts for nematode species of both medical and veterinary importance. Nematodes of the genus Angiostrongylus are of particular interest. They include Angiostrongylus cantonensis, which causes eosinophilic meningitis in humans, and the French Heartworm Angiostrongylus vasorum, a veterinary parasite that infects dogs. We have developed a molecular technique for the identification of 3rd juvenile stage A. cantonensis and A. vasorum in snail and slug intermediate hosts and are undertaking surveys to determine the incidence of A. cantonenis and A. vasorum as well as obtaining a more comprehensive understanding of nematodes that use snails as definitive and intermediate host species..
Selected Publications:
Andrus, P.S., Joof, E. & Wade, C.M. 2024. Differentiation of Bulinus senegalensis and Bulinus forskalii snails in West Africa using morphometric analysis. Acta Parasitologica. https://doi.org/10.1007/s11686-024-00830-1.
Andrus, P.S., Stothard, J.R., Kabatereine, N.B. & Wade, C.M. 2023. Comparing shell size and shape with canonical variate analysis of sympatric Biomphalaria species within Lake Albert and Victoria, Uganda. Zoological Journal of the Linnean Society 199: 713-722. https://doi.org/10.1093/zoolinnean/zlad052.
Andrus, P.S., Stothard, J.R., & Wade, C.M. 2023. Seasonal patterns of Schistosoma mansoni infection within Biomphalaria Snails at the Ugandan shorelines of Lake Albert and Lake Victoria. PLoS Neglected Tropical Diseases 17(8): e0011506.
Andrus, P. S., Rae, R. and Wade, C. M. 2022. Nematodes and trematodes associated with terrestrial gastropods in Nottingham, England. Journal of Helminthology 96: e81.
Joof, E., Sanneh, B., Sambou, S. M., Wade, C. M. 2021. Species diversity and distribution of schistosome intermediate snail hosts in The Gambia. PLoS Neglected Tropical Diseases 15: e0009823.
Joof, E., Sanyang, A. M., Camara, Y., Sey, A. P., Baldeh, I., Jah, S. L., Ceesay, S. J., Sambou, S. N., Sanyang, S., Wade, C. M. & Sanneh, B. 2021. Prevalence and risk factors of schistosomiasis among primary school children in four selected regions of The Gambia. PLoS Neglected Tropical Diseases 15: e0009380.
Standley, C.J., Goodacre, S. L., Wade, C. M. & Stothard, J. R. 2014. The population genetic structure of Biomphalaria in Lake Victoria, East Africa: Implications for Schistosome transmission. Parasites and Vectors 7: 524.
Standley, C. J., Wade, C.M. & Stothard, J. R. 2011. A fresh insight into transmission of Schistosomiasis: a misleading tale of Biomphalaria in Lake Victoria. PLoS ONE 6 (10) e26563.
Fontanilla, I. K. C. & Wade, C. M. 2008. The small subunit (SSU) ribosomal (r) RNA gene as a genetic marker for identifying infective 3rd juvenile stage Angiostrongylus cantonensis. Acta Tropica 105: 181-186.

2) Molecular Phylogenetics: Evolutionary Relationships and Taxonomy of Terrestrial and Aquatic Snails and Slugs
Snails and slugs are major components of many ecosystems, they include the vectors of many serious diseases, are important crop pests and are useful for studies on the mechanisms of evolution. We use DNA and protein sequences to build phylogenetic trees to investigate the evolutionary and taxonomic relationships in the snails and slugs, from examining the deep-level divisions in the stylommatophoran land snails and slugs, to examining the taxonomic and evolutionary relationships in the major land snail and slug groups including the Orthurethra, Helicoidea and Achatinoidea, the aquatic snails of the Hygrophila and the Caenogastropoda.
As well as our work on molecular phylogeny we are also examining population-level genetic variation within the land snails Achatina fulica, Cepaea hortensis, and Helix aspersa.
Achatina fulica: The giant African land snail Achatina (=Lissachatina) fulica originated in East Africa but is now distributed across the tropics. It is an intermediate host for the nematode parasite Angiostrongylus cantonensis, (which causes eosinophilic meningitis in humans) and is a major crop pest species. We are using genetic data to track the migration of A. fulica out of Africa and the possible link between the spread of A. fulica and A. cantonensis.

Our recent research on the introduction of African land snails into Kerala, India with Keerthy Vijayan of the Kerala Forest Research Institute can be read about here:
https://www.thehindu.com/news/national/kerala/study-finds-new-genetic-strain-of-giant-african-snail-in-kerala-and-the-uae/article66045502.ece
https://www.thehindu.com/news/national/kerala/study-finds-repeated-waves-of-giant-african-snail-invasion/article33258253.ece
Cepaea hortensis: The white lipped banded snail Cepaea hortensis is distributed across Europe, and is also found in Iceland, Greenland, as well as down the east coast of Canada and the USA. This unusual distribution pattern has led to much speculation concerning how C. hortensis got to North America and it has been suggested that the Vikings may have played a part in their introduction. We are using genetic data to examine variation in C. hortensis across continental Europe, its spread to Great Britain and Ireland and its subsequent migration to Iceland, Greenland and North America. So far our data reveal that a single genetic type exists in North America and Iceland. Interestingly, this genetic type is also present in Scandinavia, suggesting that the Vikings may indeed have played a part in the transfer of C. hortensis to Iceland and the New World.

Helix aspersa: The common garden snail Helix aspersa (=Cantareus aspersus) is another European species that is now distributed across the globe. We are examining genetic variation in H. aspersa populations to track the movements of this land snail around the world.

Selected Publications:
Zhang, G., Naggs, F., Andrus, P.S. & Wade, C.M. 2024. Phylogenetic insights into the terrestrial snails Helicoidei (Gastropoda: Stylommatophora) with special emphasis on the Camaenidae. Zoological Journal of the Linnean Society XX: 1-12. https://doi.org/10.1093/zoolinnean/zlae027.
Zhang, G. & Wade, C.M. 2023. Molecular phylogeny and morphological evolution of the Chinese land snail Cathaica Möllendorff, 1884 (Eupulmonata: Camaenidae) in Shandong Province, China. Biological Journal of the Linnean Society 140: 556-557. https://doi.org/10.1093/biolinnean/blad067.
Vijayan, K., Suganthasakthivel, R., Naggs, F., Fontanilla, I. K, Singh Soorae, P., Sajeev, T. V. & Wade, C. M. 2022. Fine-scale geographical sampling and molecular characterisation of the giant African land snail in its invasive range in Asia shows low genetic diversity, new haplotypes and the emergence of another haplotype from the Indian Ocean Islands. Biological Journal of the Linnean Society 137: 421-433. https://doi.org/10.1093/biolinnean/blac106.
Saadi, A. J., Mordan, P. B. & Wade, C. M. 2021. Molecular phylogeny of the Orthurethra (Panpulmonata: Stylommatophora). Zoological Journal of the Linnean Society 193:1126-1140.
Saadi, A.J., Davison, A. & Wade, C.M. 2020. Molecular Phylogeny of the Freshwater Snails and Limpets (Panpulmonata: Hygrophila). Zoological Journal of the Linnean Society 190: 518-531.
Saadi, A.J. & Wade, C.M. 2019. Resolving the Basal Divisions in the Stylommatophoran Land Snails and Slugs with Special Emphasis on the Position of the Scolodontidae. Molecular Phylogenetics and Evolution 139: 106529.
Nantarat, N., Sutcharit, C., Tongkerd, P., Wade, C. M., Naggs, F. & Panha, S. 2019. Phylogenetics and species delimitations of the operculated land snail Cyclophorus volvulus (Gastropoda: Cyclophoridae) reveal cryptic diversity and new species in Thailand. Scientific Reports 9: 1-12.
Fontanilla, I. K. C., Naggs, F. & Wade, C. M. 2017. Molecular phylogeny of the Achatinoidea (Mollusca: Gastropoda). Molecular Phylogenetics and Evolution 114:382-385.
Nantarat, N., Wade, C. M., Jerratthitikul, E., Sutcharit, C. & Panha, S., 2014. Molecular evidence for cryptic speciation in the Cyclophorus fulguratus (Pfeiffer, 1854) species complex (Caenogastropoda: Cyclophoridae) with description of new species. PLoS ONE 9: e109785.
Fontanilla, I. K. C., Sta. Maria, I. M. P., Garcia, J. R. M., Ghate, H., Naggs, F. & Wade, C. M. 2014. Restricted genetic variation in populations of Achatina (Lissachatina) fulica outside of East Africa and the Indian Ocean points to the Indian Ocean Islands as the earliest known common source. PLoS ONE 9: e105151.
Wade, C. M., Hudelot, C., Davison, A., Naggs, F. & Mordan, P. B. 2007. Molecular phylogeny of the helicoid land snails (Pulmonata: Stylommatophora: Helicoidea), with special emphasis on the Camaenidae. Journal of Molluscan Studies 73: 411-415.
Wade, C. M., Mordan, P. B. & Naggs, F. 2006. Evolutionary relationships among the Pulmonate land snails and slugs (Pulmonata, Stylommatophora). Biological Journal of the Linnean Society 87: 593-610.
Wade, C. M., Mordan, P. B. & Clarke, B. C. 2001. A phylogeny of the land snails (Gastropoda: Pulmonata). Proceedings of the Royal Society (London): Biological Sciences 268: 413-422.

3) Marine Evolution and Ecology: Mapping the Distribution of Genetic Variation in Marine Taxa

The foraminifera are an important group of unicellular protists that include both planktonic and benthic species. Their fossils form a high-resolution archive of past climate that plays a vital role in the campaign to understand the processes of climate change.
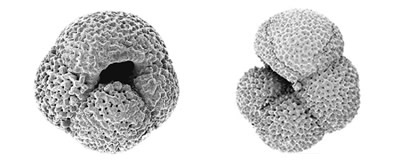
We are undertaking molecular phylogenetic studies of the foraminifera to determine evolutionary relationships within the group. Our analyses indicate that the evolutionary transition between a benthic and a planktonic way of life appears has occurred several times with the planktonic spinose species (foraminiferans with spines) clustering separately from the planktonic non-spinose species.

Our analyses show that individual mophospecies of foraminifera show an exceptionally high level of genetic diversity in their SSU rRNA genes, and many include more than one genetically distinct entity. Indeed, some of these genetic types may warrant classification as separate 'cryptic' species. This finding is of interest because of the role of foraminiferal microfossils in reconstructing past climates. For climate reconstruction it has been assumed that each 'morphospecies' is a single entity with a specific ecological (and thus climatic) preference. If the distinct genetic types within a morphospecies are in fact adapted to different habitats, and exhibit different ecological and climate preferences, then the assumption that each morphospecies is characteristic of a particular climate would be wrong. If so, there could be significant errors in the current models of climate reconstruction. Recent work suggests that different genetic types are indeed associated with different environments. If it does become possible to distinguish these newly recognised genetic types in the fossil record, the role of the foraminifera as indicators of past climate could be greatly enhanced.

Genetic studies of the foraminifera have also begun to illuminate the processes of diversification and speciation in the oceans. Despite the high degree of genetic diversity observed in their SSU rRNA genes, we have found identical sequence types (genotypes) in individuals collected at opposite ends of the globe in several morphospecies. Perhaps most remarkable is our discovery of identical rRNA genotypes in individuals collected from the Arctic and Antarctic subpolar regions within each of the cool-water morphospecies Globigerina bulloides, Turboratalita quinqueloba and Neogloboquadrina. This is surprising, as these morphospecies are only found in the high latitudes and are absent from the tropical regions, which are considered a formidable barrier to gene flow. Similarly, we have found identical rRNA genotypes within the warm-water morphospecies Orbulina universa and Globigerinoides sacculifer in individuals collected from the Caribbean and Coral Seas, and also in individuals from the Eastern Pacific and Mediterranean in O. universa. These findings are important because they suggest that gene flow can occur on a global scale within the foraminifera, with genetic intermixing between populations as far apart as the Arctic and Antarctic, or the Pacific and Atlantic. Nevertheless, our genetic data also reveals the existence of clear barriers to gene flow in the oceans. For example, genotypes of the planktonic foraminiferan Neogloboquadrina pachyderma do not transit the shallow Bering Sea with one genetic type found in the Arctic Ocean and North Atlantic, and a different genotype in the North Pacific.

As well as our work on the foraminifera we are also investigating genetic variation, gene flow and phylogeography of fish species. We have recently undertaken a population genetic study of the bluenose warehou in Tristan de Cuhna showing that fish stocks are repopulated over huge distances. We are currently investigating invasive fish species that have moved through the Suez canal to colonise the Mediterannean.
Selected Publications:
Griffiths, M., Wade, C.M., D'Agostino, D., Berumen, M.L., Burt, J.A., DiBattista, J.D. & Feary, D.A. 2024. Phylogeography of a commercially important reef fish, Lutjanus ehrenbergii, from the coastal waters of the Arabian Peninsula. Biological Journal of the Linnean Society XX: 1-12. https://doi.org/10.1093/biolinnean/blad170.
Heyworth, S. M., Bell, J. B., Wade, C. M., Cavalcante, G., Robinson, N., Young, E., Glass, J. & Feary, D. A. 2021. Genetic connectivity of seamount populations of bluenose warehou (Hyperoglyphe antartica). Frontiers in Marine Science 8: 640504. https://doi.org/10.3389/fmars.2021.640504.
Darling, K. F., Wade, C. M., Siccha, M., Trommer, G., Schulz, H., Abdolalipour, S. & Kurasawa, A. 2017. Genetic diversity and ecology of the planktonic foraminifers Globigerina bulloides, Turborotalita quinqueloba and Neogloboquadrina pachyderma off the Oman margin during the late SW Monsoon. Marine Micropaleontology 137: 64-77.
Saad, S.A. & Wade, C. M. 2017. Seasonal and spatial variation in biodiversity in saltmarsh benthic foraminiferal communities from North Norfolk, England. Microbial Ecology 73: 539.
Saad, S.A. & Wade, C. M. 2016. Biogeographic distribution and habitat association of Ammonia genetic variants around the coastline of Great Britain. Marine Micropaleontology 124: 54-62.
Seears, H. A., Darling, K. F. & Wade, C. M. 2012 Ecological partitioning and diversity in tropical planktonic foraminifera. BMC Evolutionary Biology 12: 54.
Darling, K. F., Kucera, M. & Wade, C. M. 2007. Global molecular phylogeography reveals persistent Arctic circumpolar isolation in a marine planktonic protist. Proceedings of the National Academy of Sciences (USA) 104: 5002-5007.
Darling, K. F., Kucera, M., Pudsey, C. J., & Wade, C. M. 2004. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proceedings of the National Academy of Sciences (USA) 101: 7657-7662.
Darling, K. F., Wade, C. M., Stewart, I. A., Kroon, D., Dingle, R. & Leigh Brown, A. J. 2000. Molecular evidence for genetic mixing of Arctic and Antarctic subpolar populations of planktonic foraminifers. Nature 405:43-47.

4) Bioinformatics
We are using bioinformatics tools to look at how differences in evolutionary rate between organisms can mislead phylogeny estimation (long branch attraction). We are also using bioinformatics techniques to examine questions in developmental biology and in particular we are interested in the evolutionary history of the nodal gene that has an important role in left/right asymmetry.
Selected Publications:
Davison, A., McDowell, G. S., Holden, J. M., Johnson, H. F., Koutsovoulos, G. D., Liu, M. M., Hulpiau, P., Van Roy, F., Wade, C. M., Banerjee, R., Yang, F., Chiba, S., Davey, J. W., Jackson, D. J., Levin, M. & Blaxter, M. L. 2016. Formin is associated with left-right asymmetry in the pond snail and the frog. Current Biology 26: 654-660.
Evans, T, Wade, C. M., Chapman F. A., Johnson, A. D. & Loose, M. 2014. Acquisition of germ plasm accelerates vertebrate evolution. Science 344(6180): 200-203
Selected Publications
JOOF, EBRIMA, SANNEH, BAKARY, SAMBOU, SANA M. and WADE, CHRISTOPHER M., 2021. Species diversity and distribution of schistosome intermediate snail hosts in The Gambia PLOS NEGLECTED TROPICAL DISEASES. 15(10), SAADI, AHMED J., DAVISON, ANGUS and WADE, CHRISTOPHER M., 2020. Molecular phylogeny of freshwater snails and limpets (Panpu monata: Hygrophila) ZOOLOGICAL JOURNAL OF THE LINNEAN SOCIETY. 190(2), 518-531