Structural Biology
The structural biology laboratory utilises a variety of techniques including Protein Crystallography, Cryo-electron Microscopy and associated techniques such as Isothermal Titration Calorimetry, which are then combined with functional studies or serve as a basis for structure-based drug design.
We utilise protein crystallography to investigate molecular structure and analyse protein-ligand and drug-bound complexes.
Recent breakthroughs in new cameras and powerful computational approaches for utilising Cryo-electron microscopy (Cryo-EM) have enabled us to determine, at atomic resolution, the architectures of larger molecular machines.We are also using pioneering novel chemical structural biology approaches to target proteins with small molecules or peptides.
Therapeutic areas of interest are:
- cardiovascular disease
- cancer
- neurodegeneration
- infectious diseases.
A main research focus area are proteases, an important family of proteins. Although proteases were originally studied as the molecular scissors of nonspecific protein catabolism, our view of proteases has considerably expanded. Now, protease research is key to understanding biological processes as diverse as brain function, respiratory biology, infection and immunity, cardiovascular function, cell growth and death.
The protease family represents 2% of all genes present in the human genome and is at the forefront of discovering new medicines. Examples being currently studied are serine proteases from the blood plasma such as Coagulation Factor XI, FXII and plasma kallikrein, metalloproteinases such as ADAMTS13 and cysteine proteases such as the majority of deubiquitinating enzymes.
Dr Jonas Emsley's research
3D structures of human and microbial proteins and understanding the molecular basis of human disease broadly spanning three central themes of studying:
- proteases
- cell receptors
- microbial proteins.
The crosstalk between coagulation and inflammation and has been proposed as a unifying principle for a variety of disorders as diverse as hereditary angiodema, diabetic macular edema, desquamating skin diseases and neurodegenerative disorders stroke and alzheimers.
Very few protein complex structures have been described to define the protein:protein interactions and complexes that control these so called thrombo-inflammatory pathways. I aim to utilise the results from structural studies to investigate the molecular mechanisms of thromboinflammation.
A major focus is the structure of serine and cysteine proteases to elucidate the mechanism of substrate recognition, selectivity and co-factor/membrane lipid interactions and inhibitor design.
My group has published the first structure of coagulation factor XI protease in complex with a peptide from the co-factor (Blood 2016) and in isolation (Nat Struct Mol Biol 2006).
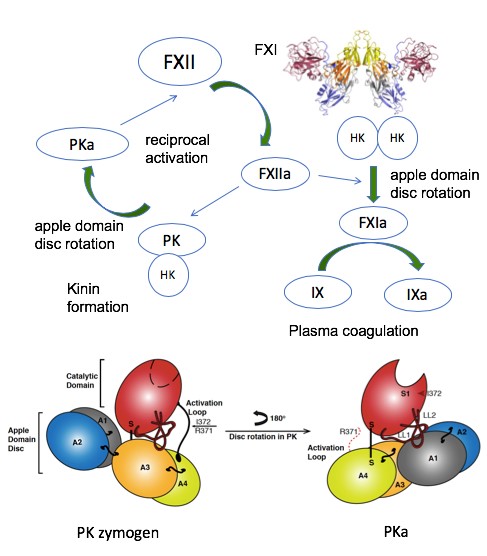
The group has recently solved the structure of the active conformation of the related apple domain containing protease prekallikrein (JTH 2019- figure below) and was involved in determining the structure of a complement protease (Science 2007).
Also the first crystal structure of the FXII protease (JTH 2015) revealed a zymogen like conformation and more recently the active form in complex with peptidomimetic inhibitor (Acta Cryst 2019) and was involved in modelling the structure of the single chain FXII active site (Blood 2017).
We have also determined the structure of the ADAMTS13 MDTCS protease domains in complex with a Fab revealing an allosteric activation mechanism (Nature Comms 2019)
Dr Ingrid Dreveny's research
Protein structure-function relationships in health and disease, enzyme specificity, protein engineering and drug discovery revolving around the areas of:
- proteases in the ubiquitin system
- posttranslational modification enzymes
- signalling proteins
- proteins in biotechnology

Tagging substrate proteins with the small modifier ubiquitin serves as an important signal for the majority of cellular processes. Deregulation of the ubiquitin-proteasome system has been implicated in the pathogenesis of many human diseases, including cancer, neurodegenerative disorders and infection.
There are 56 USPs encoded in the human genome. The Dreveny lab aims to gain a better understanding of how:
- these important cysteine proteases recognize substrates
- USPs are regulated
- they are directed into different cellular pathways
- their function can be modulated for therapeutic purposes