Contact
Biography
Helen Miranda Knight studied Natural Science as an undergraduate and completed a MRes in Neuroscience at Edinburgh University working in the in the labs of Douglas Blackwood and Richard G Morris. She was subsequently involved with the writing of 'The Hippocampus Book' working with Richard. She completed her PhD in Psychiatric Genetics at the Institute of Genetics and Cancer, University of Edinburgh. She then held a postdoctoral position under the mentorship of Trevor Robbins and Barbara Sahakian at the Behavioural and Clinical Neuroscience Institute, University of Cambridge. This was followed by an MRC Career Development Fellowship with Chris Ponting at the MRC Functional Genomics Unit (now part of the Department of Physiology, Anatomy and Genetics), University of Oxford.
In 2013, she joined the University of Nottingham to establish her own research group. Her lab investigates genomic, epigenetic, and epitranscriptomic mechanisms, with a focus on both fundamental and pathological aspects of cell biology. Current research centers on RNA metabolism and epitranscriptomic processes that contribute to synaptic function, protein aggregation, mitochondrial and mitoribosome pathways and brain diseases.
Dr Knight is the University of Nottingham co-lead for the Alzheimer's Society funded 'Lewy Body Dementia' Doctorial Training programme (Institutional lead Newcastle University). https://www.alzheimers.org.uk/what-we-do/our-research/alzheimers-society-doctoral-training-centres/lewy-body-dementia
She was an elected member of the Senate since 2020-2023 and a member of the UoN Knowledge Exchange Committee from 2021-2023. Dr Knight is also a Genetic Society Ambassador https://genetics.org.uk/
Expertise Summary
My research interests span neurodevelopmental disorders, including intellectual disabilities, schizophrenia, and psychosis, as well as neurodegenerative conditions such as Lewy body diseases (e.g. dementia with Lewy bodies and Parkinson's disease dementia, Parkinson's disease), and mitochondrial disorders. We employ a range of molecular techniques to investigate genomic, transcriptomic, and proteomic profiles, aiming to uncover novel molecular features that are either fundamental to all cell types or unique to specialised cells such as neurons. We also use these omics' techniques and large datasets, advanced microscopy (STEM, TEM, SRM, confocal), neuronal cell and iPSC cultures, and human brain tissue, to examine how disease pathology develops and progresses.
In recent years, my team has focused on the emerging field of RNA modifications, also known as epitranscriptomics, and their role in regulating cellular function. We have investigated key RNA modifications such as N6-methyladenosine (m6A), 5-methylcytosine (m5C), RNA capping modifications, and other less common forms of RNA methylation. We are interested in RNA binding proteins especially the RNA methylation protein effector machinery which control RNA modification dynamics and molecular interactions within sub-cellular compartments and cell nanodomains. We are trying to understand how these processes contribute to, and regulate, change in transcriptional and translational outputs, synaptic and neuronal function, protein aggregation and granule disassembly.
Teaching Summary
My teaching interests are in human molecular and medical genetics, epigenetics and disease pathology of brain and neurological disorders, as well as diagnostics and molecular services in health care.… read more
Research Summary
My lab focuses on investigating the epigenetic basis of cellular function and how these processes may be disrupted in brain diseases and cognitive traits. We employ a range of molecular techniques to… read more
Selected Publications
FLITTON, MILES, RIELLY, NICHOLAS, WARMAN, RHIAN, WARDEN, DONALD, SMITH, A. DAVID, MACDONALD, IAN A. and KNIGHT, HELEN MIRANDA, 2019. Interaction of nutrition and genetics via DNMT3L-mediated DNA methylation determines cognitive decline NEUROBIOLOGY OF AGING. 78, 64-73 KNIGHT, H.M., PICKARD, B.S., MACLEAN, A., MALLOY, M.P., SOARES, D.C., MCRAE, A.F., CONDIE, A., WHITE, A., HAWKINS, W., MCGHEE, K., VAN BECK, M., MACINTYRE, D.J., STARR, J.M., DEARY, I.J., VISSCHER, P.M., PORTEOUS, D.J., CANNON, R.E., ST CLAIR, D., MUIR ,W.J. and BLACKWOOD, D.H.R., 2009. A cytogenetic abnormality and rare coding variants identify ABCA13 as a candidate gene in schizophrenia, bipolar disorder, and depression American Journal of Human Genetics. 85(6), 833-846 PICKARD, B.S., KNIGHT, H.M., HAMILTON, R.S., SOARES, D.C., WALKER, R., BOYD, J.K.F., MACHELL, J., MACLEAN, A., MCGHEE, K.A., CONDIE, A., PORTEOUS, D.J., ST. CLAIR, D., DAVIS, I., BLACKWOOD, D.H.R. and MUIR, W.J., 2008. A common variant in the 3'UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder Proceedings of the National Academy of Sciences. 105(39), 14940-14945
Current Research
My lab focuses on investigating the epigenetic basis of cellular function and how these processes may be disrupted in brain diseases and cognitive traits. We employ a range of molecular techniques to investigate genomic, transcriptomic, and proteomic profiles, aiming to uncover novel molecular features that are either fundamental to all cell types or unique to specialised cells such as neurons. We also use these 'omics' techniques including direct RNA Oxford Nanopore sequencing, as well as large datasets, advanced microscopy (STEM, TEM, SRM, confocal), neuronal cell and iPSC cultures, and human brain tissue, to examine how disease pathology develops and progresses. The different strands of lab include:
1. RNA epi-transcriptomics, brain function and the dark proteome
We have demonstrated in vitro and in situ brain tissue that m6A modified RNAs are in pre- and post- synaptic sites. We have also shown that the m6A eraser, ALKBH5, is involved in active translation during synaptic plasticity; and that the YTHDF1 and YTHFDF3 readers exhibit differential roles during synaptic maturation. We are currently further characterising the role of m6A, m5C and rarer forms of RNA modifications in synaptic plasticity, protein aggregation including alpha synuclein mechanisms, ribosome regulation and translational outputs, and mitochondria processes.
m6A-sequencing of human parahippocampus brain tissue and neuronal cell cultures revealed distinct white and grey matter m6A methylome profiles, but in both neuronal and glial cell-rich tissue, indicated that m6A and m5C effector proteins are themselves highly modified. We have also shown that RNA modification systems cross talk with posttranslational modification processes and with each other to regulate distinct cellular processes.
We are also investigating new roles for enzymes involved in RNA metabolism and how translation of non-conventional proteins such as non-conical open reading frames (ncORFs) and small proteins known as microproteins are regulated. We are identifying and characterizing novel proteins, referred to as the 'dark proteome' by using ribosequencing, RNA sequencing and mass Spectrometry.
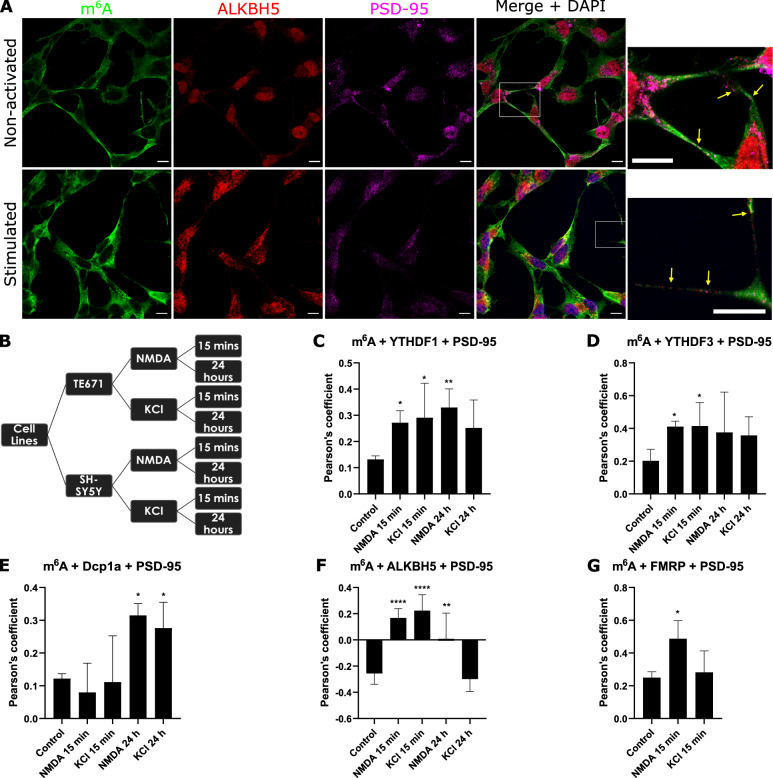
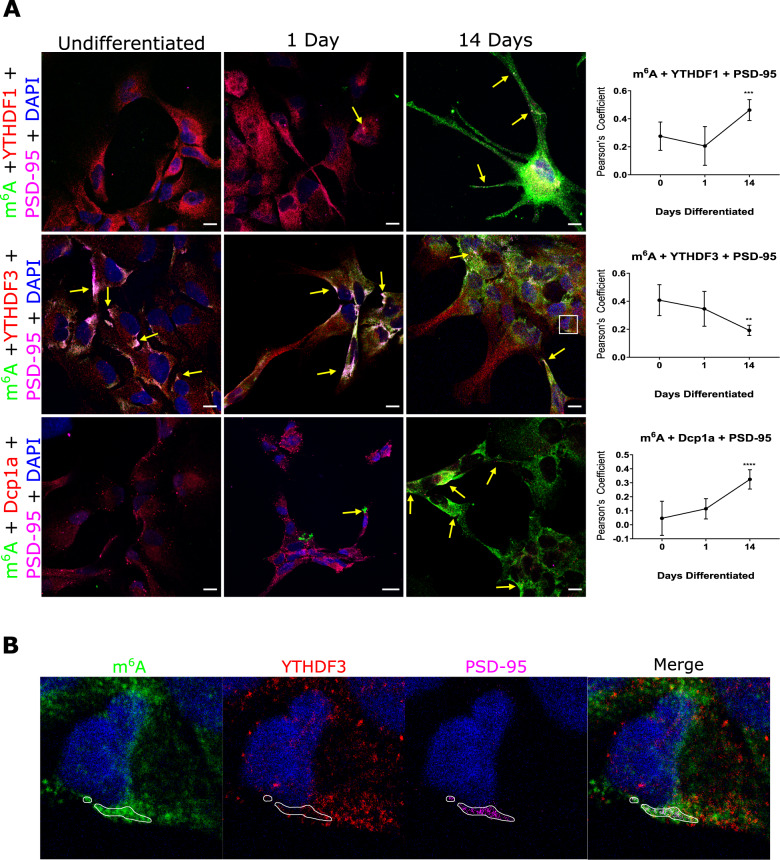
m6A-modified RNA and the demethyltransferase ALKBH5 increase in abundance during synaptic plasticity and synaptic maturation during development.
Current lab members: Joseph Stones; Oliver Orji; Hala Shaker.
Past Lab members: Braulio Martinez De La Cruz; Eleanor Bellows, Maria Isabel Haig, Merve Demirbugen Öz.
Papers:
Martinez De La Cruz, B., Markus, R., Malla, S. et al. …Knight, HM., Modifying the m6A brain methylome by ALKBH5-mediated demethylation: a new contender for synaptic tagging. Mol Psychiatry 26, 7141-7153 (2021). https://doi.org/10.1038/s41380-021-01282-z
Baron F, Zhang M, Archer N, Bellows E, Knight HM, Welham S, Rutland CS, Mongan NP, Hayes CJ, Fray RG, Bodi Z. The importance of m6A topology in chicken embryo mRNA: a precise mapping of m6A at the conserved chicken β-actin zipcode. RNA. 2023 Jun;29(6):777-789. doi: 10.1261/rna.079615.123.
Bellows, E., Fray, R.G., Knight, H.M., and Archer, N. (2024). The expanding role of cap-adjacent modifications in animals. Frontiers in RNA Research 2. 10.3389/frnar.2024.1485307.
2. RNA epi-transcriptomics and brain disease
We have discovered that the abundance of different forms of modified-RNA and effector protein machinery are disrupted in neurodegenerative diseases and that in some diseases, such a Dementia with Lewy body, there is significant reduction in modified-RNAs and location of modified RNAs within neuronal cells, e.g. Parkinson's disease (PD). This suggests that although there may be similar pathological features across diseases, e.g., alpha synuclein aggregates, disruption to RNA metabolism processes maybe a driver of pathology and molecular differences between the diseases.
We have also identified changes in RNA expression and protein abundance in m5C writer and readers effector protein in Alzheimer's disease (AD) and traumatic brain injury, and in m6A reader and anti-reader effector proteins in AD and MCI brain tissue.
In collaboration with clinicians affiliated with the Children's Brain Tumour Centre, Nottingham, we are examining how methylated RNAs and transcriptional control may be contributing to rare pediatric tumour aetiology.

m5C RNA effector proteins are differentially expressed in Alzheimer's Disease and Traumatic brain Injury.

m6A-modified RNA is significantly more abundant in Lewy body dementia and significantly reduced and misplaced brain cells in Parkinson's Disease (PD) across human brain regions.
Lab members: Adriana Perez Grovas-Saltijeral, Hala Shaker, Lauryn Walker, Masar Radhi.
Past Lab members: Braulio Martinez De La Cruz; Merve Demirbugen Öz, Eleanor Bellows, Maria Isabel Haig.
Papers:
Martinez De La Cruz, B., …Knight, H.M., "m6A mRNA methylation in human brain is disrupted in Lewy body disorders", Neuropathol Appl Neurobiol. 2023; 49(1).
Perez Grovas-Saltijeral, A., …Knight, H.M., 'Differential Expression of m5C RNA methyltransferase genes NSUN6 and NSUN7 in Alzheimer's disease and Traumatic Brain Injury', Mol Neurobiol. 2023 Apr;60(4):2223-2235.
Knight,H. M., Demirbugen Öz. M., PerezGrovas-Saltijeral, A.,. Dysregulation of RNA modification systems in clinical populations with neurocognitive disorders. Neural Regeneration Research :10.4103/1673-5374.385858.
Orji OC, Stones J, Rajani S, Markus R, Öz MD, Knight HM. Global Co-regulatory Cross Talk Between m6A and m5C RNA Methylation Systems Coordinate Cellular Responses and Brain Disease Pathways. Mol Neurobiol. 2024 Nov 5. doi: 10.1007/s12035-024-04555-0.
PerezGrovas-Saltijeral, A., Stones, J., Orji, O. C., Shaker, H., and Knight, H. M. (2025). 'Modification of the RNA methylome in neurodevelopmental disorders'. Current Opinion in Genetics and Development, 92, Article102330. DOI: 10.1016/j.gde.2025.102330. Invited review article.
- Biomarkers for early Alzheimer's disease, Dementia with Lewy Bodies, cognitive decline and psychosis
In collaboration with clinical colleagues, we are using brain imaging and molecular analysis to explore biomarkers for changes in cognitive performance and symptoms in psychosis. We are also identifying novel extracellular vesicles transcriptional changes as biomarkers for dementia with Lewy bodies and Parkinson's disease.
Current and past Lab members: Gamze Gizem Tekeli, Fatma Busra Isik
Papers:
Isik FB, Knight HM, Rajkumar AP. Extracellular vesicle microRNA-mediated transcriptional regulation may contribute to dementia with Lewy bodies molecular pathology. Acta Neuropsychiatr. 2023 Jun 21:1-10. doi: 10.1017/neu.2023.27.
Past Research
1. Rare coding variants and genetic architecture of complex disease
ABCA13 ATP-binding cassette sub-family A member 13
A patient with schizophrenia was found to have a chromosomal abnormality which directly disrupted one gene, ABCA13, a lipid membrane transporter protein. Through exon sequencing of the functional domains of ABCA13, multiple rare coding variants were identified. Follow-up pedigree and case-control association studies strongly suggest that mutations within this gene, significantly contribute to the genetic aetiology of schizophrenia, bipolar disorder and major depression. This work raises key questions into the type of mutations underlying complex disease, for example; the frequency of rare risk variants (private to a family, ultra-rare, or 1-2%), whether carrying 2 or more rare coding variants (compound heterozygotes) is a feature of risk burden, and how common it is to have variants that contribute to several disorders which cross diagnostic categories. ABCA13 is now known to be involved in vesicle cholesterol accumulation and synaptic vesicle endocytosis and has been confirmed as a genetic risk gene for psychiatric disorders.
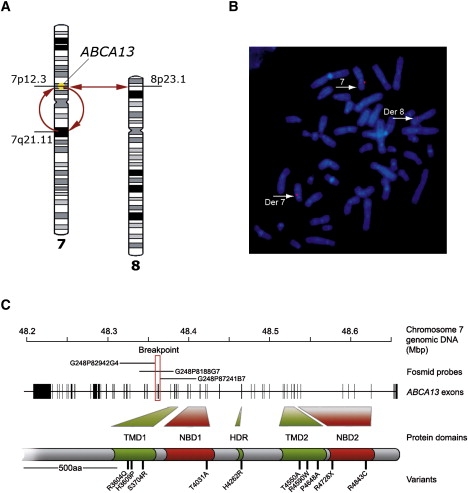
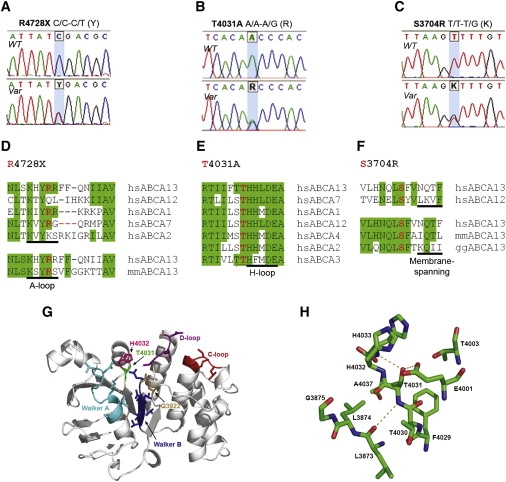
ABCA13 cytogenetic lesion and rare coding variants in families with Schizophrenia, bipolar disorder and major depression
Knight, H.M., Pickard, B.S., et al.,…Blackwood, D.H (2009) "A cytogenetic abnormality and rare coding variants identify a transporter as a candidate gene for schizophrenia, bipolar disorder and depression". The American Journal of Human Genetics, 85:1-14.
2. Glutamatergic neurotransmitter genes and risk for neurodevelopmental disorders
GRIK4/GLUK4 glutamate ionotropic kainite receptor 4 subunit
A deletion within the 3' untranslated region of the GRIK4 gene was identified as a protective factor for bipolar disorder. RNA secondary structure analysis in conjunction with evidence of increased relative mRNA abundance of the protective deletion allele suggested that this common variant may have a functional impact through differential mRNA stability. Using immunohistochemistry, GLUK4 protein expression was characterized in key regions of human brain and a genotype/protein expression correlation study indicated that GLUK4 expression was significantly increased in subjects carrying the protective allele. The protective deletion allele is also associated with better cognitive performance.



A 'protective' deletion within GRIK4/GLUK4 is increased in the hippocampus and cortex and associated with higher cognitive performance.
Past Lab members: Maria Koromina, Miles Flitton.
Papers:
Koromina, M., Flitton. M., UK10K., Mellor, I., Knight, H.M. (2019) "A Kainate receptor GluK4 deletion, protective against bipolar disorder, is associated with enhanced cognitive performance across diagnoses in the TwinsUK cohort". World Journal of Biological Psychiatry, 20(5):393-401.
Koromina. M., Flitton. M., Blockley. A., Mellor. I.R., Knight, H.M. (2019) "Damaging coding variants within kainate receptor channel genes are enriched in individuals with schizophrenia, autism and intellectual disabilities". Sci Rep.16;9 (1):19215.
Knight HM, Walker R, James R, Blackwood DH, Muir WJ, Pickard BS. (2012) GRIK4/KA1 protein expression in human brain tissue and correlation with a genetic variant associated with bipolar disorder. Am J Med Genet B Neuropsychiatr Genet. 159B(1):21-9.
Pickard BS, Knight HM, Hamilton RS, Soares DC, Walker R, Boyd JK, Machell J, Maclean A, McGhee KA, Condie A, Porteous DJ, St Clair D, Davis I, Blackwood DH, Muir WJ (2008) A common variant in the 3'UTR of the GRIK4 glutamate receptor gene affects transcript abundance and protects against bipolar disorder. PNAS 105:14940-14945.
3. Functional Genomics, DNA methylation and methionine metabolism factors
Disease risk alleles and non-coding functional effects.
Genome-wide array case-control association studies and population/pedigree next generation re-sequencing studies frequently give rise to results implicating regions or variants which are not in coding regions of genes but are potentially associated/linked with a trait or disease. How do we interpret these non-coding signals? And how do we tell if common variants are directly contributing to a phenotype or are merely associated through being near a causative variant (synthetic association)? One method currently used is to examine whether the variants are located within or/and directly disrupt regulatory element and thus could have a functional non-coding effect. This involves bioinformatic data mining of cohort and functional genomic datasets for regulatory features such as different species of non-coding RNAs, DNA and RNA methylation, as well as functional studies.

Cognitive change correlates with methionine metabolism factors and DNMT3L genotype.
Past lab members: Nicholas Rielly, Miles Flitton.
Papers:
Flitton, M., Rielly, N., Warman, R., Warden, D., Smith, A D., Macdonald, I.A., Knight, H.M. (2019) "Interaction of nutrition and genetics via DNMT3L-mediated DNA methylation determines cognitive decline". Neurobiol Aging. Jun;78:64-73.
Flitton. M., Macdonald, I.A., Knight, H.M. (2019) "Vitamin intake is associated with improved visuospatial and verbal semantic memory in middle-aged individuals". Nutritional Neuroscience, 22(6):401-408. Epub 2017.
Davies,G., Harris, S.E., Reynolds, C.H., Payton, A., Knight, H.M., Liewald, D.C., et al., (2014) A Genome-Wide Association Study implicates the APOE/TOMM40 locus in non-pathological cognitive ageing. Molecular Psychiatry, 19(1):76-87.