Datasets and Tools
Tool: SuRVoS
SuRVoS brings together machine learning models, computer vision techniques and human knowledge within a user interface to interactively segment large 3D volumes.
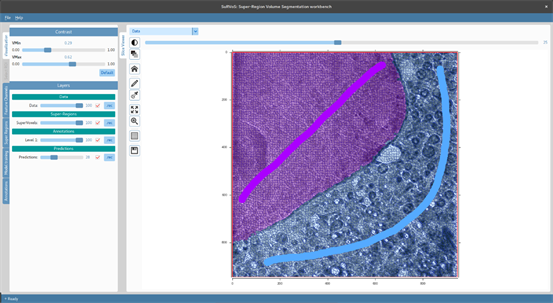
Website - https://diamondlightsource.github.io/SuRVoS/
Dataset and Tools: RootSystemML
RootSystemML is a file format to represent root architectural data. It has been designed to overcome two major challenges:
- To enable portability of root architecture data between different software tools in an easy and interoperable manner allowing seamless collaborative work,
- To provide a standard format upon which to base central repositories which will soon arise following the expanding worldwide root phenotyping effort.
RSML allows to store 2D or 3D image metadata, plant and root properties and geometries, continuous functions along individual root paths and a suite of annotations at the image, plant or root scales, at one or several time points. The plant ontologies are used to describe botanical entities that are relevant at the scale of root system architecture.

Website - https://rootsystemml.github.io/
CVL People - Michael Pound
Dataset and Tools: CellSet
CellSeT is a tool to segment and analyse confocal microscope images.
CellSeT can be used to analyse confocal images of plant cells. Image analysis algorithms are used to extract the network structure of the visible cells. This information is then used to perform quantitative analysis of the cell structure, and guide additional analysis through the use of an extensible plug-in system, which allows processes to be applied to individual and groups of cells. Plug-ins have been written for a variety of tasks, e.g. estimating the amount of a marker protein in a cell. CellSeT is written in C# .NET, and as such runs only on Windows machines with an up-to-date version of the .NET framework installed. Plug-ins can be written in any .NET compatible language, such as C# or VB. For users who do not have access to a Windows machine, we have successfully used CellSeT in a virtual windows environment.
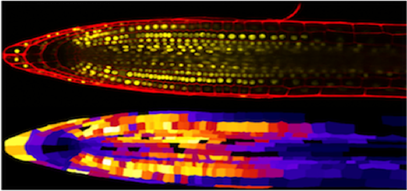
Website - Cellset
CVL People - Michael Pound
Dataset and Tools: Canopy Reconstruction
Accurate and useful models of plants in three dimensions, based on multiple colour images using a low-cost camera.
Our reconstruction work aims to produce accurate and useful models of plants in three dimensions, based on multiple colour images using a low-cost camera. Beginning with an initial point cloud, obtained using standard structure from motion tools, the scene is first filtered, and segmented into small patches that represent sections of leaf surface. Each surface is individually represented by a best-fit plane, and the boundary of each surface is adjusted to better fit the image and modelling data. This adjustment is achieved using the level set method, which optimises the smoothness of each surface, while growing or shrinking them in response to image information and neighbouring surfaces.

Website - Canopy Reconstruction
CVL People - Andrew French
CVL People - Michael Pound
Dataset and Tools: RootNav
RootNav is a software tool for quantification of root system architectures in a range of plant species, grown and imaged in a variety of ways.
The Expectation-Maximisation algorithm is used to classify the initial image, and produce a probability map of regions likely to contain root material. An intuitive user interface allows the user to guide in the detection of complex root paths, including the ability to overcome areas of challenging root structure, or image noise. Measurements can be exported directly, or saved in a database, where additional traits can be calculated later using a simple plug-in system. RootNav is written in C# .NET, and as such runs only on Windows machines with an up-to-date version of the .NET framework installed. It is possible to run RootNav on a Mac computer using software such as parallels.
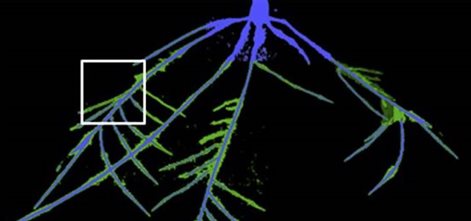
Website - RootNav
CVL people - Michael Pound
Dataset and Tools: Acid
Annotated Crop Image Database

Website - https://plantimages.nottingham.ac.uk/