Machine Learning for Automated Quality Control
Supervisors: Matteo Bastiani, Stam Sotiropoulos, Rob Dineen
Research Proposal
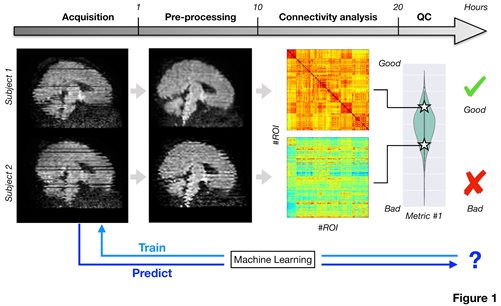
Brain connectivity can be probed in vivo and non-invasively using functional and diffusion MRI. However, resultscan be affected by factors related neither to physiology, anatomy or pathology; inevitable hardware and subjectrelatedartefacts and distortions due to, e.g., excessive motion [1], can adversely affect connectivity analyses.Therefore, automated quality control (QC) [2] is of great importance, especially in vulnerable population studies,such as neonates [3] and children affected by different pathologies. Moreover, the extraction of quality metrics canhelp to standardise data from different smaller studies. This would allow to spread the costs and difficultiesassociated with patient recruiting and to speed up the development of imaging-based biomarkers.We have recently developed novel QC pipelines and processing tools, which boost significantly our ability toextract useful information from challenging datasets. However, these pipelines are computationally-demanding.The main aim of this project is to develop complementary machine learning techniques to automatically identify andextract relevant QC features from several available large cohort early developmental studies. These features willthen be used both to predict the quality of newly acquired datasets prior to any pre-processing step and to allowcross-site data standardisation (Fig. 1). The available large cohort datasets will include different data acquisitionprotocols and will sample the early developmental period, from close to birth [3] to early childhood [4]. Datasets canbe also augmented including other protocols, site and pathologies sampling from the ADHD200 [5] and ABIDE Iand I cohorts [6]. This will provide a comprehensive set of normal and pathological developmental scenarios to trainthe automated machine learning models and will ensure the generalizability of the proposed approaches. Thespecific aims include:- Extract relevant QC metrics after pre-processing to automatically classify the overall dataset quality: What doesmake a good dataset? Is it the average signal-to-noise ratio (SNR) or the average amount of motion? Usingmachine learning techniques, the student will train automated models capable of classifying multi-modal datasetsbased on their QC metrics. Moreover, careful feature extraction and ranking will allow to learn which of thosefeatures play a more important role in such classification.- Learn suitability of a dataset for connectivity analysis from the raw data and automatically identify datasets thatare not fit for purpose: Based on the framework developed under the previous aim, the student will developmethods to automatically evaluate the quality of a dataset from raw un-processed data. Crucially, this will allow toavoid expensive and lengthy pre-processing pipelines and could inform clinicians or experimenters on whichdataset need to be reacquired, if possible.- Develop ways that allow comparable across scanners and across studies results for multi-modal connectivitydata: Pooling datasets from different studies can be very effective to study different developmental diseases.However, differences in acquisition protocols, scanner hardware and image reconstruction algorithms can lead toapparent results which are not reflecting any true biological change. Using the models trained before, the studentwill develop automated methods to standardise datasets across sites and scanners to ensure comparable results.
A medium-term goal of the NIHR Nottingham BRC is ‘To develop advanced motion correction, image analysis and statistical tools to facilitate efficient and cost-effective clinical application. (RA4)’. Our approach of providing a device for evaluating motion minimisation acquisition techniques will facilitate this goal and is complementary to approaches that optimise post-processing of images to reduce motion artefacts
References: [1] Andersson, J., & Sotiropoulos, S., 2016, NeuroImage [2] Bastiani, M., et al., 2019, NeuroImage [3]Bastiani, M., et al., 2018, NeuroImage [4] Dineen, R., et al., 2017, Proc. ISMRM [5] Consortium, H.D., et al., 2012,Front Syst Neurosci [6] Di Martino, A., et al., 2017, Sci Data