Liquid Chromatography Mass Spectrometry (LC-MS)
LC-MS enables the quantitative and qualitative analysis of small compounds
Liquid Chromatography-Mass Spectrometry
LC-MS is a versatile and highly sensitive analytical technique for the measurement of small molecular weight compounds in a diverse range of sample types. It uses a series of mass detection systems to provide both quantitative and qualitative analyses. Situated in the Centre for Analytical Bioscience within the School of Pharmacy our MS facilities are ideal for small molecule analysis (<2000m/z) and are complemented by a suite of UHPLC or specialised surface analysis interfaces (DESI, LESA) and software for data interpretation.
Capabilities
- Ultra-high-performance liquid chromatography (UHPLC) coupled to either high resolution-MS or QTrap MS.
- Accurate mass measurement (<5 ppm) for untargeted metabolomics and high resolution targeted quantitative analysis.
- Multiple reaction monitoring (MRM) and parallel reaction monitoring (PRM) for high sensitivity or specific quantitative analysis.
- Multi-stage MS and data-dependant MS fragmentation for structural elucidation and MS/MS identification.
- Ambient analysis of chemical compounds on biological/material surfaces using LESA-MS.
- Direct infusion high throughput analysis with NanoMate (no LC separation).
- Spot analysis or imaging with AP-MALDI (5 µm resolution).
Typical Applications
- Quantification of drug/drug metabolites, endogenous metabolites/lipids and contaminants in cells, microorganisms, plants, biofluids, tissues and environmental samples.
- Global metabolite profiling, metabolomics, biomarker discovery and surface analysis.
- Understanding the metabolic effects of biotic and/or abiotic perturbations on a biological system, for example drug-induced changes to intracellular metabolite pathways.
- Absolute quantification of a wide range of intra and extracellular metabolites using isotope-dilution mass spectrometry.
- Isotope-assisted metabolic pathway profiling.

How does LC-MS work?
Liquid chromatography (LC) provides a universal separation of chemical components in solution-based mixtures. It does so by the different affinities of the chemical compounds with a liquid mobile phase flowing through a stationary phase.
Mass spectrometry (MS) involves the ionisation of chemical species, followed by the detection of the mass-to-charge ratio (m/z) and intensity of the various ionised analytes. In the case of LC-MS this is following a chromatographic separation.
The huge complexity of the metabolites in biological systems has led to LC-MS being utilised to reduce sample complexity by separating out the various sample components prior to their MS detection. Therefore, LC-MS is a powerful analytical technique not only for the quantification of small biomolecules (metabolites) but also for the identification of unknown and known compounds in biological samples.
While traditionally a bulk analysis, modern technical adaptations usually pertaining to the ionisation method, allow LC-MS variants to be used as a powerful complimentary surface analytical tool.
Selected Publications
1. Metabolic alterations in dairy cattle with lameness revealed by untargeted metabolomics of dried milk spots using direct infusion-tandem mass spectrometry
To determine whether metabolic signatures associated with lameness could be discovered with untargeted metabolomics, we developed a novel workflow using direct infusion-tandem mass spectrometry to rapidly analyse (2 min per sample) dried milk spots (DMS) that were stored on commercially available Whatman® FTA® DMPK cards for a prolonged period (8 and 16 days). An orthogonal partial least squares-discriminant analysis (OPLS-DA) method validated by triangulation of multiple machine learning (ML) models and stability selection was employed to reliably identify important discriminative metabolites. With this approach, we were able to differentiate between lame and healthy cows based on a set of lipid molecules and several small metabolites.
Wenshi He, Ana S. Cardoso, Robert M. Hyde, Martin J. Green, David J. Scurr, Rian L. Griffiths, Laura V. Randall and Dong-Hyun Kim, Analyst, 2022, 147, 5537
2. Liquid Extraction Surface Analysis-Mass Spectrometry Using Superhydrophobic-Superhydrophilic Patterning
The use of the Droplet Microarray (DMA) provides a surface-assisted LESA-MS method delivering significant improvement of the surface extraction repeatability leading to the acquisition of more robust and higher quality data. Such a method shows potential to be used for LESA-MS for controlled and reproducible surface extraction and for acquisition of high quality, qualitative data in a high-throughput manner.
Meurs J, Alexander MR, Levkin PA, Widmaier S, Bunch J, Barrett DA and Kim D-H, Anal. Chem. 2018, 90, 10, 6001-6005
3. Probing Interkingdom Signaling Molecules via Liquid Extraction Surface Analysis–Mass Spectrometry
Here, we present a model system for growing biofilms on discs before utilizing rapid and direct surface sampling MS, namely, liquid extraction surface analysis, to study the microbial exometabolome. One of the benefits of this approach is its surface-specific nature, enabling mimicking biofilm formation in a way that the study of planktonic liquid cultures cannot imitate. Our model system provides a route to investigate changes in the exometabolome, such as metabolites that become circulatory in the presence of multiple pathogens, and provides a rapid analytical approach to gaining a mechanistic understanding of bacterial signalling.
Shaun N. Robertson, Fadi Soukarieh, Thomas M. White, Miguel Camara, Manuel Romero, and Rian L. Griffiths, Anal. Chem. 2023, 95, 11, 5079–5086
4. Quantitative Bioreactor Monitoring of Intracellular Bacterial Metabolites in Clostridiumautoethanogenum Using Liquid Chromatography–Isotope Dilution Mass Spectrometry
We report a liquid chromatography–isotope dilution mass spectrometry method for the simultaneous quantification of 131 intracellular bacterial metabolites of Clostridium autoethanogenum. A comprehensive mixture of uniformly 13C-labeled internal standards (U-13C IS) was biosynthesized from the closely related bacterium Clostridium pasteurianum using 4% 13C–glucose as a carbon source. The U-13C IS mixture combined with 12C authentic standards was used to validate the linearity, precision, accuracy, repeatability, limits of detection, and quantification for each metabolite. The method was subsequently applied for the daily monitoring of the intracellular metabolites of C. autoethanogenum during a CO gas fermentation over 40 days as part of a study to optimize biofuel production. The concentrations of the metabolites were estimated at steady states of different pH levels using the robust-fitting mathematical approach, and we demonstrate improved accuracy of results compared to conventional regression.
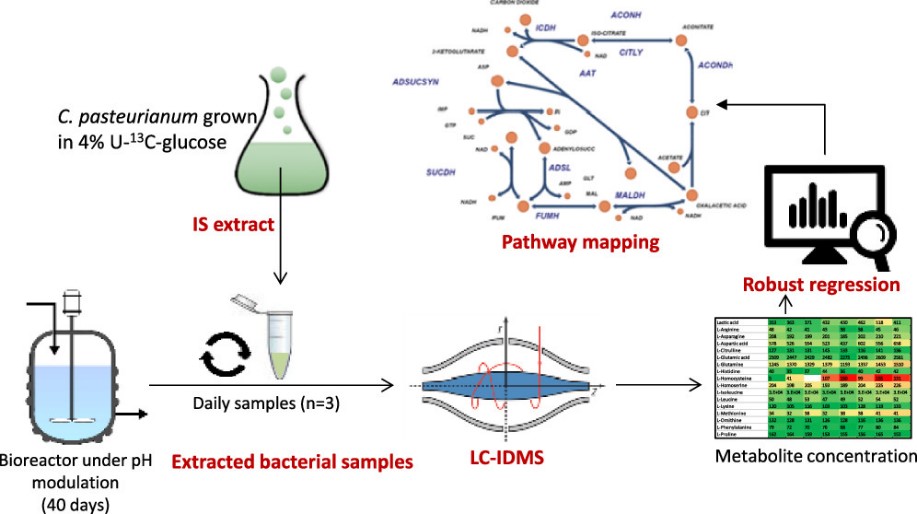
Laudina Safo, Salah Abdelrazig, Alexander Grosse-Honebrink, Thomas Millat, Anne M. Henstra, Rupert Norman, Neil R. Thomas, Klaus Winzer, Nigel P. Minton, Dong-Hyun Kim, and David A. Barrett, ACS Omega 2021, 6, 21, 13518–13526
5. Advantages of using biologically generated 13C-labelled multiple internal standards for stable isotope-assisted LC-MS-based lipidomics
In this study, an in vivo13C labelling strategy was employed to explore four species (Escherichia coli, Arthrospira platensis, Saccharomyces cerevisiae and Pichia pastoris) as a source of 13C-labelled internal standards (13C-ISs) for more accurate and quantitative liquid chromatography (LC)-mass spectrometry (MS)-based lipidomics. Overall, use of a biologically generated 13C-IS lipid mixture of 357 identified lipid ions resulted in significant reduction in the lipid CV% of normalisation compared with other normalisation methods using total ion counts or a commercially available deuterated internal standard mixture. This improved normalisation using 13C-IS was confirmed in a typical lipidomics analysis using a large number of samples (>100+) and long analysis time (>70 h).
Malak A. Jaber, Bruna de Falco, Salah Abdelrazig, Catharine A. Ortori, David A. Barrett and Dong-Hyun Kim, Anal. Methods, 2023, 15, 2925
6. Metabolic characterisation of THP-1 macrophage polarisation using LC–MS-based metabolite profiling
This study represents successful application of LC–MS metabolomics approach to characterise M1 and M2 macrophages providing functional readouts that show unique metabolic signature for each phenotype. This data could contribute to a better understanding of M1 and M2 functional properties and could pave the way for developing new therapeutics targeting different immune diseases.
Alaa Abuawad, Chidimma Mbadugha, Amir M. Ghaemmaghami & Dong-Hyun Kim, Metabolomics, 2020, 16:33
Our LC-MS Facilities
QTRAP 6500+ and QTrap 4000 Quadrupole Linear Ion Trap LC-MS/MS
- Equipped with HPLC and UHPLC.
- Quantitative and qualitative targeted metabolite profiling, and for ID confirmation. Used where sensitivity is required.
- Advanced MS scanning options and information dependant acquisition offer unique approaches to metabolite profiling and identification.
Thermo Fisher QExactive (Hybrid quadropole Orbitrap) high resolution mass spectrometer
- Equipped with NanoLC, UHPLC and TriVersa NanoMate..
- Small molecules profiling and identification.
- Quantitative and qualitative measurements.
- Metabolomics and lipidomics with high resolution measurements.
Thermo Orbitrap Fusion Lumos Tribrid Mass Spectrometer
- Resolution > 500,000 FWHM.
- MSn capabilities.
TriVersa NanoMate, Liquid Extraction Surface Analysis-Mass Spectrometry (LESA-MS)
- Surface extraction and analysis under ambient conditions.
- Direct infusion high throughput analysis.
- Advantages where UHV analysis may not be suitable; alternative to ToF-SIMS and MALDI.
- Coupled to QExactive MS to achieve a high resolution and confidence in identification using data dependant MS/MS.
- The new LESAPLUS allows for automated LESA experiments plus additional nano-LC separation.
Ambient Pressure Matrix Assisted Laser Desorption Ionisation (AP-MALDI)
- Surface analysis/imaging under ambient conditions.
- Unlike regular MALDI, it does not require a vacuum chamber, which means the analysis is faster and the samples can be kept in their native statuss.
Data Analysis
- We perform data analysis for untargeted metabolomics with Compound Discoverer 3.3 (Thermo Scientific) and multivariate analysis with SIMCAP.
- For quantitative analysis we use TraceFinder (Thermo Fisher Scientific), Analyst ( Sciex) and MultiQuant.